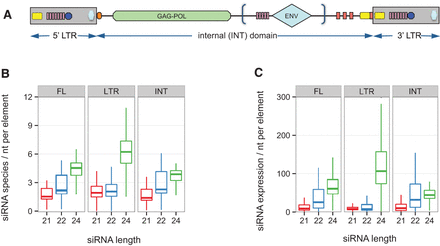
The Sirevirus genome and general siRNA mapping patterns. (A) A schematic of a Sirevirus element (based on Bousios et al. 2010). The gag/pol genes are shown in green, and the envelope-like gene (present in some families only) as a light blue diamond. An inverted repeat (IR; yellow) surrounds the junction of the internal (INT) domain with the 3′ LTR. The outmost 5′ side of the junction is occupied by a polypurine-tract (PPT; red). Additional PPTs cluster upstream of the junction. The palindromes (pink) are located in the cis-regulatory area of the LTRs preceding a conserved TATA box (blue circle), and near the envelope-like gene (when present). The 5′ LTR/INT domain junction harbors a C-rich integrase signal (light blue hexagon) and the primer binding site (orange box). (B,C) Number of siRNA species (B) and expression (C) per nucleotide of the full-length (FL), LTR, or INT domain of all 6456 Sireviruses based on the leaf library.











