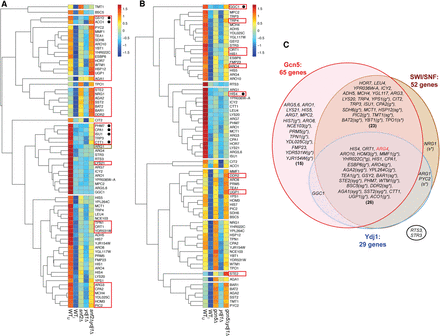
Snf2, Gcn5, and Ydj1 cooperate in promoter H3 eviction at many of the 70 exemplar induced genes. (A,B) Hierarchical clustering analysis of the average relative H3 occupancies in the [−1, NDR, +1] regions was performed in R using the “pheatmap” function, with the following parameters: scale = “row,” clustering_method = “average,” clustering_distance_rows = “euclidean,” cluster_rows = TRUE, cluster_cols = FALSE, cutree_row = 10. The avg. H3 occ. values for each gene given in Supplemental Figures S13 and S14 were transformed to z-scores. Boxed genes display additive effects of mutations; those with bullets show strong eviction defects only in double mutants. (C) Venn diagram depicting involvement of Snf2, Gcn5, or Ydj1 at the 70 exemplar genes deduced from effects of single, double, or triple mutations eliminating or depleting these cofactors on H3 promoter occupancies in SM-treated cells, quantified in Supplemental Figures S13–S15 and summarized in Supplemental Figure S18. (g*, s*, or y*) Strong defect in H3 eviction in gcn5Δ, snf2Δ, or ydj1Δ single mutants, respectively, with H3 occupancies similar to those in WTU cells. As described in Supplemental Results, there was generally good agreement between conclusions reached from ChIP-seq (Supplemental Figs. S13–S16) and conventional ChIP (Supplemental Fig. S3B) for ARG1, ARG4, HIS4, and CPA2. However, the lower variance of conventional ChIP data for ARG4 (Supplemental Fig. S4A) led us to place it among the group of genes employing all three cofactors.











