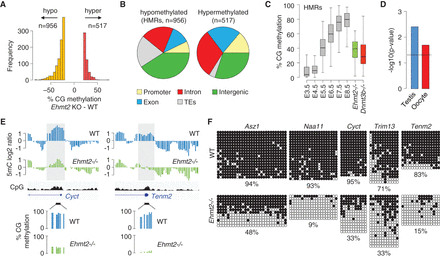
Identification of hypomethylated regions (HMRs) in Ehmt2−/− embryos. (A) Histogram of the number of sequences that gain or lose >20% methylation in the RRBS from Ehmt2−/− compared with WT embryos. (B) Pie charts representing the percentages of hypomethylated and hypermethylated sequences mapping to promoters, gene bodies, intergenic regions, and transposable elements (TEs). (C) Boxplot showing the dynamics of methylation at HMRs (filtered to lose >30% methylation in Ehmt2−/− embryos) in early embryos, Ehmt2−/− E8.5 embryos (green), and Dnmt3b−/− E8.5 embryos (red). (D) Preferential site of expression of genes with a promoter-proximal HMR (−1000 to +1000 bp relative to the TSS). The dashed line represents the position for a P-value of 0.05. (E) Examples of promoter-proximal HMRs. The sites of hypomethylation identified by MeDIP (gray boxes) confirm the differences measured by RRBS. (F) Bisulfite cloning and sequencing of selected promoter HMRs in WT and Ehmt2−/− E9.5 embryos. The values below the sequencing data indicate the percentage of methylated CpGs in the amplicon.











