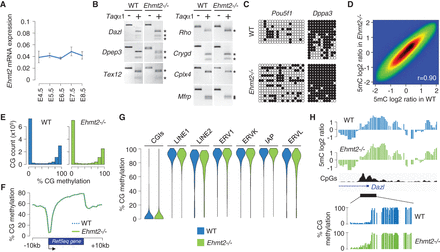
Impact of EHMT2 on the methylome in mouse embryos. (A) Expression of Ehmt2 mRNAs measured by RT-qPCR in early mouse embryos, depicted as a ratio relative to the expression of two housekeeping genes (Actb and Rpl13a; mean ± SEM, n = 2 technical replicates on five to 10 pooled embryos). (B) Promoter DNA methylation of candidate genes measured by COBRA in WT and Ehmt2−/− E9.5 embryos. The restriction fragments marked with an asterisk are end products of the digestion (indicating DNA methylation). (C) Promoter DNA methylation of the Pou5f1 and Dppa3 genes measured by bisulfite sequencing in WT and Ehmt2−/− E9.5 embryos. Circles represent methylated (black) or unmethylated (white) CpG dinucleotides; each horizontal line is one sequenced clone. (D) Pairwise comparison of methylated DNA immunoprecipitation (MeDIP) log2 ratios at individual oligos in WT and Ehmt2−/− E9.5 embryos. The density of data points increases from blue to dark red. The Pearson correlation coefficient (r) is indicated on the graph. (E) Density histograms of RRBS CpG methylation scores in WT and Ehmt2−/− E8.5 embryos. (F) Average distribution of RRBS methylation over RefSeq genes and flanking sequences in WT and Ehmt2−/− embryos. (G) Violin plots of RRBS methylation measured in CpG islands (CGIs) and retrotransposons. (H) Example of methylation profiles in the promoter of the Dazl gene in WT and Ehmt2−/− embryos. The upper tracks depict smoothed MeDIP log2 ratios of individual oligonucleotides, and the bottom tracks depict RRBS methylation scores at individual CpGs. The CpG density is shown in black.











