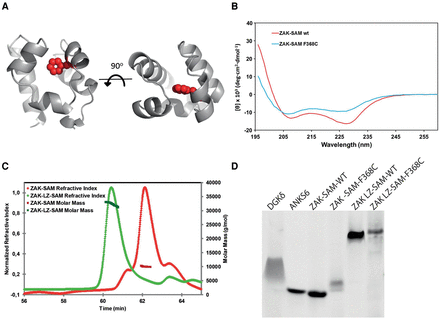
Functional effects of ZAK SAM domain missense mutation. (A) Phyre model of the ZAK SAM domain based on the structure of the SAM domain of DGKD (30% identity, PDB ID 3BQ7). Phe368 resides in the hydrophobic core of the SAM domain and is shown as spheres highlighted in red. (B) Circular dichroism spectra of wild-type ZAK-SAM and the Phe368Cys mutant. The altered spectrum of ZAK-SAM Phe368Cys correlates with a loss of alpha-helicity. (C) Purified ZAK-SAM and ZAK-LZ-SAM were assessed by SEC-MALS. The SAM domain alone is monomeric with an observed molecular weight (MW) of 10.2 kDa (predicted MW = 8.9 kDa). The LZ-SAM construct has an observed MW of 32.3 kDa, corresponding to a homogenous population of dimer (predicted dimer MW = 29.6 kDa). (D) negGFP fusions of wild-type and Phe368Cys mutants of ZAK-SAM and ZAK-LZ-SAM assessed by native gel electrophoresis. In this assay, described previously (Liu et al. 2000), proteins are fused to a highly negative green fluorescent protein (negGFP). The negative charges on negGFP cause reliable migration toward the cathode in a native gel, and the relative mobility reflects oligomerization. Monomeric SAM domains migrate as discrete bands and with higher mobility than polymeric SAM domains, which migrate slower and with a smeared character. negGFP fusions of a monomeric SAM (ANKS6) and a polymeric SAM (DGKD) are shown as controls. ZAK-SAM appears monomeric, and addition of the LZ causes a significant gel shift. The Phe368Cys (F368C) substitution in the ZAK SAM domain causes a retarded migration, indicating protein aggregation.











