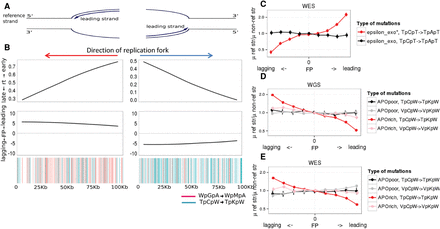
APOBEC mutational signature as a function of fork polarity in cancer genomes. (A) Diagram explaining the correspondence between the reference strand being replicated as leading or lagging and the direction of the replication fork. (B) Characteristics of the 50 genomic regions of lengths of 100 kb each with the highest (left) and lowest (right) FP on WGS data set for APOrich tumors. Horizontal axis: coordinates within the considered genomic regions. (Upper) average of the replication times (RT) across these 50 regions; (middle) fork polarity values reconstructed as derivatives of RT represented in the upper panel; FP > 0 corresponds to the leading strand, and FP < 0 corresponds to the lagging strand; (bottom) color-coded APOBEC signature mutations on reference strand within these regions. Each vertical line corresponds to a mutation. (C,D,E) The ratio of the mutation rates of the considered mutation type to its reverse compliment on the reference strand as a function of the propensity of the replication fork to replicate the reference strand as lagging or leading. Horizontal axis: genome split by FP values into nine bins from low FP (bin 1) to high FP (bin 9). Vertical bars represent 95% confidence intervals. (C) Pol ε_exo* produces mutations on the leading strand; APOBEC causes mutations on the lagging strand in WGS (D) and WES (E) data sets.











