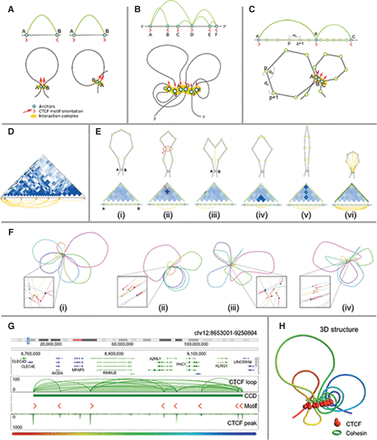
Modeling of loop structures. (A) Two possible chromatin loop structures associated with CTCF motif orientation: hairpin loop structure resulting from the convergent CTCF motifs (left), and coiled loop structure resulting from tandem CTCF motifs (right). (B) Schematic representation of a CCD unit and possible chromatin loops stretch out of an interconnected interaction complex containing all the anchors bound by CTCF. (C) Simulation model on the subloop level. Genomic distance wp between two anchors of a loop (subloop) is translated to physical distance dp between the corresponding beads. During the simulation, the angles αi between two flanking segments (lines) are also considered. CTCF motif orientation could force the chromatin fiber to form different shapes of loops for convergent (A,B) and tandem (B,C) orientations. (D) An example of a subloop structure indicated by a heat map constructed by using all intra-loop PET singletons, with the heat maps generated assuming simple polymer physics. (E) Potential impacts of the intra-loop PET singleton data onto the likely shapes (i–vi) of chromatin loop. (F) Effect of including different subloop level energy terms onto the shapes of chromatin loops presented on a selected interaction block (Chr 17: 63226521–64565170). A zoomed-in view on the interaction complex is presented for each structure (spheres denote the anchors, red lines denote the CTCF motif orientations). (F[i]) In the base model, only loop segment lengths and angles between them are considered, resulting in regularly shaped and freely distributed loops. (F[ii]) Including the CTCF motif orientation in the model introduces more constraints and thus results in limited mobility of loops, which become aligned to each other. (F[iii]) Considering the subanchor heat map introduces more constraints on the loops’ shape and results in loops being more irregular. (F[iv]) Finally, including both CTCF motif orientation and subanchor heat maps yields irregular but aligned loops. For clarity, the simulation was restricted to two dimensions. (G) An example CCD identified at Chr 12: 8665991–9229876, with several anchors with well-defined CTCF motif orientations. (H) Predicted 3D structure for the CCD shown in G. A highly organized CTCF core is surrounded by chromatin loops of various sizes.











