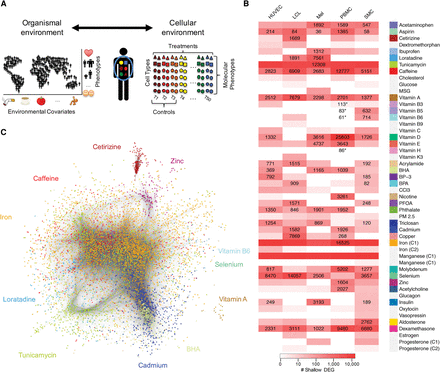
Overview of gene expression response. (A) Schematic of experimental design and rationale. Our approach uses specific treatment conditions as tightly controlled proxy for the organism environment and measures molecular phenotypes, such as gene expression, to infer genetic and molecular mechanisms for complex traits. (B) Heatmap of differential gene expression. Shown for each cell type (columns) and treatment (rows) combination are the number of differentially expressed genes (10% FDR and |log2FC| > 0.25). The shade of red indicates the number of differentially expressed genes from an initial screening step (see Supplemental Texts 5 and 8.1). Cellular environments with a strong response were further sequenced to a higher depth (>58 M reads, 113 M on average), and the number of differentially expressed genes is indicated by the text. Environments marked with an asterisk were chosen to confirm that treatments with a small response from the shallow sequencing data similarly have a small response when deep sequenced. Colors next to treatment names represent treatments chosen for deep sequencing. Gray indicates treatments that were not deep sequenced. (C) Global coexpression network inferred using weighted gene correlation network analysis (WGCNA) on 14,535 genes. Each dot represents a gene. Each module is assigned a color based on the treatment with the highest eigengene t-value. Note that colors representing treatments are consistently used across all the figures.











