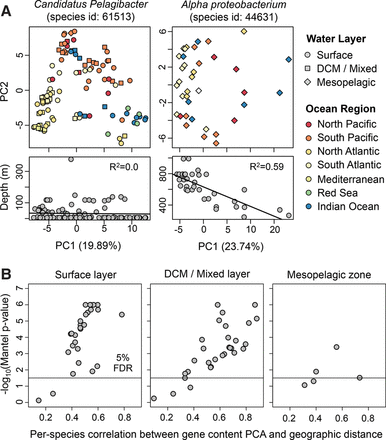
Gene content and geography are correlated for many marine bacteria. (A) Principal component analysis (PCA) of gene content for two bacterial species. Each point indicates a bacterial population from a different seawater sample. Point color and shape indicate the marine region and water layer, respectively. Candidatus Pelagibacter populations tend to cluster together based on ocean region, not ocean depth. In contrast, Alpha proteobacterium populations tend to cluster together based on ocean depth, not ocean region. (B) Gene content PCA and geographic distance are significantly correlated for most prevalent marine species. PCA distance was calculated using the Euclidian distance between PC1 and PC2 of the gene presence–absence matrix. Geographic distance was calculated using the great-circle distance between sampling locations. For each species, the correlation of these two distances (horizontal axis) and associated P-value (vertical axis) were computed using the Mantel test with 1 million permutations. Only one metagenome per location was included in the tests. The population structure of marine bacteria, based on the first two principal components of gene content, is correlated with geography for many species of bacteria.











