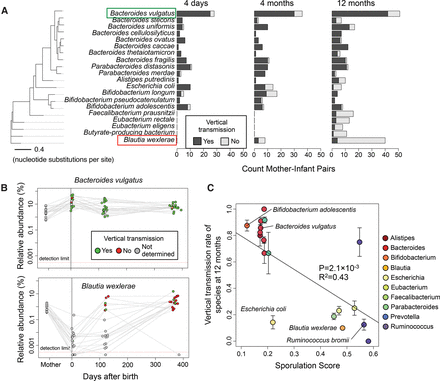
Distinct timing and vertical transmission patterns for microbiome species. (A) Vertical transmissions for bacterial species across mother–infant pairs at three time points. The 20 species with the greatest number of high-coverage mother–infant pairs are shown. A vertical transmission is defined as >5% marker allele sharing between mother and infant. The phylogenetic tree is constructed based on a concatenated DNA alignment of 30 universal genes (Supplemental Fig. S3) and shows that phylogenetically related species have similar transmission patterns. (B) Bacteroides vulgatus is an early colonizing species that is frequently transmitted vertically, whereas Blautia wexlerae is a late colonizing species that is rarely transmitted vertically. Gray points indicate there was insufficient sequencing coverage to quantify SNPs and determine transmission. (C) Species with low vertical transmission rates are predicted to be spore-formers with the ability to survive in the environment. Sporulation scores are genomic signatures of sporulation based on 66 genes (Browne et al. 2016). Error bars indicate one standard error in each direction. Only species with sporulation scores computed by Browne et al. (2016) and with three or more mother–infant pairs at 12 mo are shown.











