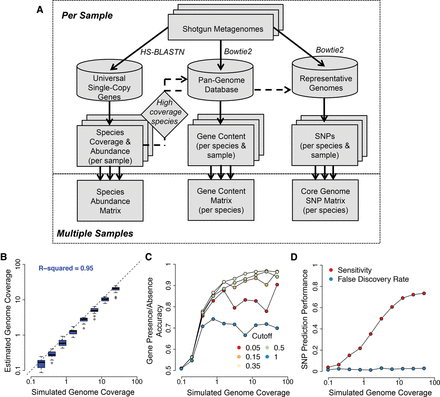
An integrated pipeline for profiling species abundance and strain-level genomic variation from metagenomes. (A) The MIDAS analysis pipeline. Reads are first aligned to a database of universal-single-copy genes to estimate species coverage and relative abundance per sample. For species with sufficient coverage, reads are next aligned to a pan-genome database of genes to estimate gene coverage, copy number, and presence–absence. Finally, reads are aligned to a representative genome database to detect SNPs in the core genome. The core genome is defined directly from the data by identifying high-coverage regions across multiple metagenomic samples. (B–D) To evaluate performance for each component of MIDAS, we analyzed 20 mock metagenomes composed of 100-bp Illumina reads from microbial genome-sequencing projects. Each community contained 20 organisms with exponentially decreasing relative abundance. We tested the ability of MIDAS to estimate species coverage and to predict genes and SNPs present in the strains of the mock communities compared to the reference gene and genome databases. (B) Species coverage is accurately estimated. Each boxplot indicates the distribution of estimated genome coverages across 20 mock communities for the top eight most abundant species out of 20 analyzed. (C) Gene presence–absence is accurately predicted when genome coverage is above 1×, and a gene copy number cutoff of 0.35 is used. Accuracy = (Sensitivity + Specificity)/2; Sensitivity = (number of genes correctly predicted as present)/(number of total genes present); Specificity = (number of genes correctly predicted as absent)/(number of total genes absent). (D) SNPs are detected with a low false-discovery rate and good sensitivity when genome coverage is above 10×. Sensitivity = (number of correctly called SNPs)/(number of total SNPs); False Discovery Rate = (number of incorrectly called SNPs)/(number of called SNPs).











