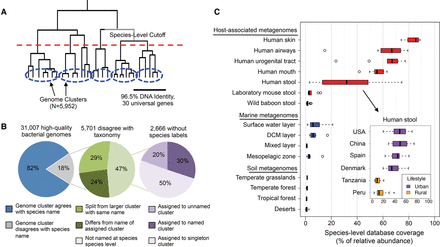
Construction of bacterial species database and its coverage of microbial communities across different environments. (A) In total, 31,007 genomes were hierarchically clustered based on the pairwise identity across a panel of 30 universal gene families. We identified 5952 species groups by applying a 96.5% nucleotide identity cutoff across universal genes, which is equivalent to 95% identity genome-wide. (B) Concordance of genome-cluster names and annotated species names. Of the 31,007 genomes assigned to a genome cluster, 5701 (18%) disagreed with the consensus PATRIC taxonomic label of the genome cluster. Most disagreements are due to genomes lacking annotation at the species level (47%). Other disagreements are because a genome was split from a larger cluster with the same name (29%) or assigned to a genome cluster with a different name (24%). (C) Coverage of the species database across metagenomes from host-associated, marine, and terrestrial environments. Coverage is defined as the percentage (0%–100%) of genomes from cellular organisms in a community that have a sequenced representative at the species level in the reference database. The inset shows the distribution of database coverage across human stool metagenomes from six countries and two host lifestyles.











