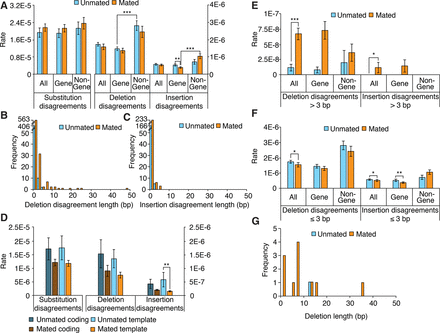
Characterization of duplex disagreements and comparison to deletion lengths. (A) Duplex disagreement rates broken down by genome location. (B,C) Distribution of duplex insertion and deletion disagreement lengths. (D) Duplex disagreement rates broken down by coding or template strand type. (E) Duplex disagreement rates broken down by genome location for disagreements longer than 3 bp. (F) Duplex disagreement rates broken down by genome location, for disagreements not exceeding 3 bp. (G) Deletion lengths. (A, D–F) Error bars, SEM; asterisks denote significance of pairwise differences computed using bootstrapped confidence intervals ([*] P < 0.05; [**] P < 0.01; [***] P < 0.001), with P-values corrected using Hochberg's step-up procedure with a 5% false-discovery rate.











