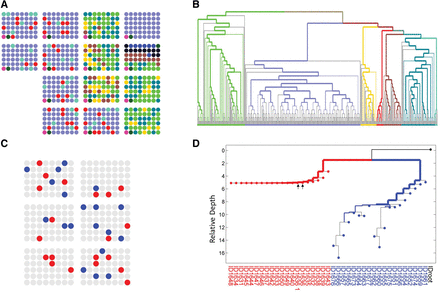
In vivo cell lineage tree reconstruction of human cells. To validate the reconstruction of human in vivo samples, we first selected single-cell samples from seven human individuals and distributed them among different AA chips. (A) Representation of different cell samples in 48-well batches (circles) in 14 AA chips, with colors indicating different source individuals. (B) As expected, cell lineage reconstruction of samples from A demonstrates accurate reconstruction of human samples in accordance with individual donors. The width of the colored branches represents the significance of the clustering, which was calculated using a hypergeometric test (wider = lower P-value) (see Supplemental Note S3). Branches are colored in accordance with the colors in A. The two bottom left samples of each AA chip correspond to a positive control (dark green) and negative control (pink). (C,D) Cell lineage reconstruction of melanoma and normal lymphocytes from the same patient (YUCLAT) (Krauthammer et al. 2015). (C) Same representation as in A: Metastatic melanoma (red) and normal PBL (blue) were randomly distributed over six AA chips. (D) Cell lineage reconstruction of samples from C demonstrates a perfect separation between the two cell populations. Arrows indicate a SC sample duplicate.











