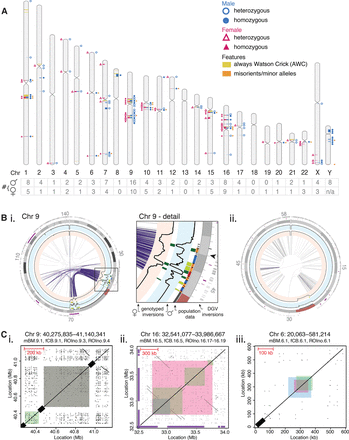
Genome-wide comparison of inversion profiles of an adult male and newborn female. (A) The inversion profile characterized for a male (blue circles, right-hand side) and female (pink triangles, left-hand side). Each inversion (plotted using Idiographica v2.2 [Kin and Ono 2007]) was genotyped as either heterozygous (empty symbols) or homozygous (filled symbols). The number of inversions per chromosome is listed below (table). The location of Always Watson Crick regions (yellow) and misorients or minor alleles (orange) are also depicted. (B) Invert.R histograms (black lines) for the adult male (blue background) and newborn female (pink background) were overlaid on Circos plots, with all inversions plotted (heterozygous in light green, homozygous in dark green). See Supplemental Data File S1 for other chromosomes. Palindromic intra-chromosomal segmental duplications (purple lines) correlate with the inversion load of each chromosome. (i) Chr 9 contains several inversions clustered within palindromic segmental duplications (purple links). Structural differences of inversions were seen between the two donors within this complex region of the genome (Chr 9, detail). The arrowhead marks the area depicted in C, i. (ii) Nonpalindromic segmental duplications (gray links) are common on Chr 19, which contains a single inversion. (C) Dot plots illustrate the genomic architecture of inversions, which can be flanked by (i) reference assembly gaps (black bars on diagonal axis), (ii) palindromic segmental duplications, or (iii) nonrepetitive sequence. Sequence coordinates that were self-aligned are listed above each plot, with the inversions found in the male (mBM; blue), female (fCB; pink), and pooled donor population (ROIno; green) highlighted. Inversions listed in the Database of Genomic Variants are shown (purple bars on x- and y-axes). See Supplemental Data File S2 for all other inversions.











