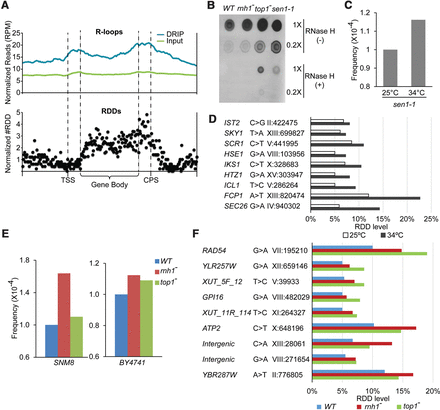
RDDs and R-loops are coupled. (A) Genome-wide distribution of R-loops and RDDs. Metagene plots show overlap between R-loops and RDDs: (TSS) transcription start site; (CPS) cleavage and polyadenylation signal; (gene body) annotated coding sequence in reference genome. (B) Nucleic acid blots probed with S9.6 antibody show more RNA–DNA hybrids in rnh1−, top1−, and sen1-1 mutants compared to wild-type control. In the temperature-sensitive mutant of sen1-1, at nonpermissive temperature, RDD frequency (C) and levels (D) are higher. (E) In two different strain backgrounds, RDD frequencies are higher in rnh1− and top1− mutants than in wild-type control. (F) RDD levels are higher in rnh1− and top1− mutants than in wild-type control. Two Xrn1-sensitive unstable transcripts (XUTs) are annotated according to XUT track in the SGD genome browser (van Dijk et al. 2011).











