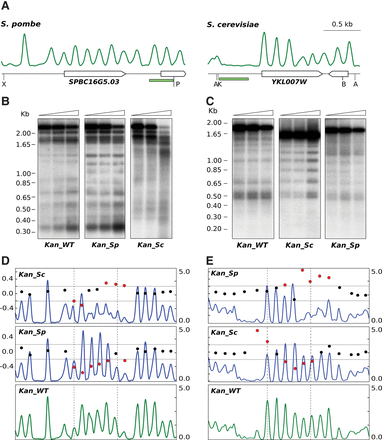
Engineering nucleosomal positioning on prokaryotic genes. (A) Nucleosomal distribution across the indicated regions in S. pombe and S. cerevisiae. Genes are represented by pointed rectangles. Restriction sites for XhoI (X), PstI (P), AvaII (A), Kpn2I (K), and BstxI (B) and the localization of the hybridization probes (green) are shown. (B) MNase end-labeling analysis of a S. pombe strain where the SPBC16G5.03 ORF has been replaced by the wild-type kanamycin ORF (Kan_WT) or a version where their codons have been replaced by the synonymous codons with the highest score in the PSWM of S. pombe (Kan_Sp). The same sequence is unable to position nucleosomes when it replaces the YKL007W ORF in the genome of S. cerevisiae (Kan_Sp in C). (C) MNase end-labeling analysis of a S. cerevisiae strain where the YKL007W ORF has been replaced by the wild-type kanamycin ORF (Kan_WT) or a version where their codons have been replaced by the synonymous codons, with the highest score in the PSWM of S. cerevisiae (Kan_Sc). The same sequence is unable to position nucleosomes when it replaces the SPBC16G5.03 ORF in the genome of S. pombe (Kan_Sc in B). (D) DANPOS analysis of modified kan ORFs in S. pombe and (E) in S. cerevisiae shows a negative score relative to the wild-type kan in each of them (middle panels). Dotted vertical lines indicate the position of the kan ORF. The positive score when the two modified sequences are swapped (top panels) indicates a lower degree of positioning than the unmodified wild-type kan ORF in the two yeasts.











