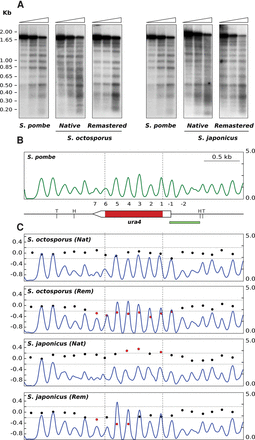
Engineering nucleosomal positioning on eukaryotic coding DNA sequences. (A) MNase end-labeling analysis of wild-type S. pombe ura4 (left panel). The pattern of internucleosomal bands becomes more diffused when the S. pombe ura4 ORF is replaced by the native S. octosporus or S. japonicus ORFs. The original sharp profile is restored when the codons of the two ORFs are replaced by synonymous codons, with the highest score in the PSWM of S. pombe (Remastered). (B) Nucleosome occupancy map of wild-type S. pombe. Restriction sites for HindIII (H) and TfiI (T) and the localization of the hybridization probe (green bar) are indicated. (C) Nucleosome occupancy maps of the strains harboring the native (Nat) and remastered (Rem) ura4 ORF of S. octosporus and S. japonicus (blue). DANPOS analysis of nucleosome positioning of the four mutant strains relative to the wild type. Symbols are as in Figure 1.











