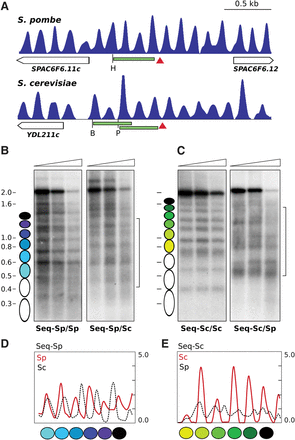
Engineering nucleosomal positioning on synthetic DNA sequences. (A) Nucleosomal profile of the genomic regions of S. pombe and S. cerevisiae, where the artificial sequences were inserted (red arrowheads). The restriction sites for HindIII (H), BsmI (B), and PagI (P) and the localization of the hybridization probes (green) are indicated. (B) The MNase end-labeling analysis of the S. pombe sequence integrated in S. pombe generates a regular nucleosomal profile as predicted (colored ovals) (Seq-Sp/Sp). Insertion of the same sequence in S. cerevisiae (bracket) generates a different pattern (Seq-Sp/Sc). (C) The S. cerevisiae sequence generates a regular profile after integration in S. cerevisiae (Seq-Sc/Sc) but fails to position nucleosomes when integrated in the S. pombe genome (Seq-Sc/Sp, bracket). (D) MNase-seq occupancy maps of the S. pombe artificial sequence integrated in S. pombe (red) or in S. cerevisiae (black). (E) MNase-seq occupancy maps of the S. cerevisiae artificial sequence integrated in S. cerevisiae (red) or in S. pombe (black).











