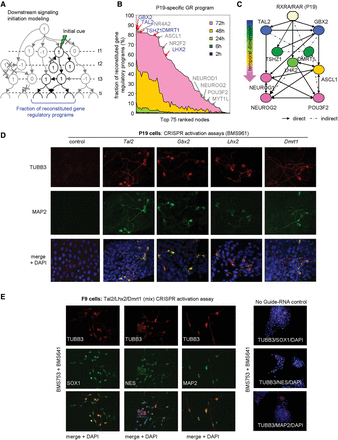
Predicting master regulators of neurogenesis by modeling signal transduction propagation. (A) Scheme of the signal transduction propagation model initiated at a downstream layer in the reconstructed GRN. (B) 1087 nodes comprising the P19-specific GRP (x-axis) ranked according to their performance in reconstituting the ultimate level of the P19-specific program (y-axis). Previously known neuronal factors are depicted in association with their position in the ranking (gray). Less characterized factors with significant signal propagation performance toward the final level are in blue. (C) Transcriptional regulatory relationships among the newly predicted factors in B are depicted in the context of their interconnections with relevant neuronal markers. Their relative temporal transcriptional response under RA-driven conditions is indicated (color coded). (D) Immunofluorescence micrographs illustrating the presence of the neuronal markers TUBB3 (red) and MAP2 (green), in P19 cells after CRISPR/dCas9 (D10/N863A)-mediated transcription activation of Tal2, Gbx2, Lhx2, or Dmrt1 treated with the RARG-specific agonist BMS961 or vehicle. (E) Immunofluorescence micrographs revealing the presence of the neuronal markers TUBB3 (red) and MAP2, SOX1, or Nestin (NES; green) in F9 cells after CRISPR/dCas9 (D10/N863A)-mediated transcription activation of Tal2, Lhx2, and Dmrt1 treated with BMS961 and the RARB-specific BMS641. In the right panel, a mock-CRISPR/dCas9 (D10/N863A) transfection assay (no guide RNA) in F9 cells under identical treatment conditions is displayed.











