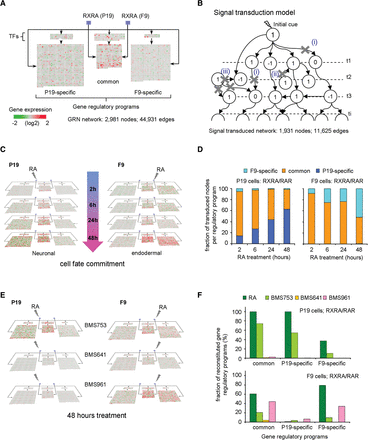
Temporal signal propagation in RA-induced GRNs for neuronal and endodermal cell fate decisions. (A) Structure of the reconstructed GRN displaying genes that are selectively or commonly regulated during neuronal and endodermal cell differentiation. For illustration purposes, all edges were removed; arrows indicate the direct regulation of each of these programs by TFs that are bona fide direct RA responsive genes (blue squares; black arrows). Gene expression changes are illustrated as heat maps. (B) Signal transduction model aiming at evaluating the coherence between the reconstructed GRN and the temporal gene expression changes. The starting node where the initial cue activates the signal transduction is depicted, as well as the downstream node interconnections required for its propagation. The temporal transcriptional state for each node is defined as 1, 0, or −1 (up-regulated, nonresponsive, or down-regulated, respectively). The model excludes signal cascade progression branches (illustrated by crosses) when (1) the state of a node remains nonresponsive; (2) the directionality of the TF-TG relationship is opposite to the temporal signal flux; or (3) the TF-TG relationships are not part of the main signal transduction propagation branches. (C) Temporal transcriptional evolution of the reconstructed GRNs in P19 or F9 RA-induced cell differentiation. Note that common programs dominate at early time points, while the neuronal/endodermal programs take over at late time points. (D) Fraction of transduced nodes per regulatory program for both model systems (F9-specific, common, P19-specific), as assessed by the signal transduction model. As illustrated in C, the common gene regulatory program is activated early (>80% in both cell lines after 2 h of RA treatment), while the cell fate-specific program is set up progressively (∼60% of specific programs in either of the model systems after 48 h of RA). (E) Responsiveness of common and neuronal-/endodermal-specific GRNs described in A to agonists selective for the three RAR isotypes. (F) Fraction of reconstituted gene regulatory programs (GRPs) (after 72 h of RA treatment) in both model systems when either the RA or RAR-specific agonists-derived transcriptomes are used for modeling signal transduction propagation.











