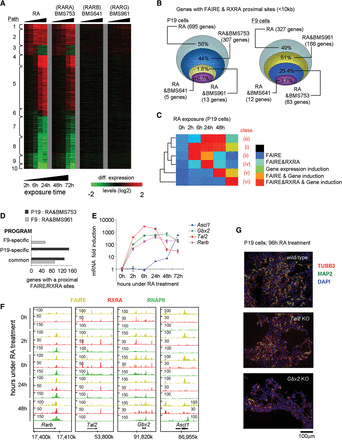
Different RAR subtypes induce chromatin alteration in RA-responsive genes of P19 and F9 cells. (A) Heat map illustrating the transcriptional responses of genes comprising the 10 coexpression paths characterized in P19 cells during RA-induced differentiation or in the presence of the indicated RAR isotype-specific agonists. (B) DEGs during RA-induced differentiation in P19 or F9 cells that present FAIRE and RXRA binding in proximity (<10 kb from the TSS) are compared with their corresponding transcriptional response in the presence of RAR isotype-specific agonists. (C) Heat map illustrating temporal SOTA classification of P19 genes positive for RXRA binding, and/or display altered chromatin structure (FAIRE-seq), and/or are induced in response to RA. This classification gave rise to the identification of six classes of genes with different temporal induction patterns (Supplemental Fig. S5). (D) Number of DEGs F9 or P19 cells commonly regulated by RA and BMS753 or RA and BMS961 and presenting a proximal FAIRE and RXRA binding site, stratified for the cell-specific (P19, F9) and common programs. (E) RT-qPCR revealing the temporal RA-induced mRNA expression profiles of bona fide RA target genes. (F) FAIRE-seq, RXRA, and RNAPII ChIP-seq profiles for the factors assessed in E. Rarb, Gbx2, and Tal2 are early responding genes, while Ascl1 gets significantly induced only after 24 h of RA induction. (G) Immunofluorescence micrograph of wild-type and CRISPR/Cas9-inactivated Tal2 or Gbx2 P19 cells after 96 h of RA treatment. Cells were stained for the neuronal markers TUBB3 (red) and MAP2 (green); nuclei were stained with DAPI (blue). Gbx2-inactivated cells present a lower frequency of double-stained TUBB3/MAP2 cells and shorter axon-like extension than Tal2-inactivated or wild-type cells.











