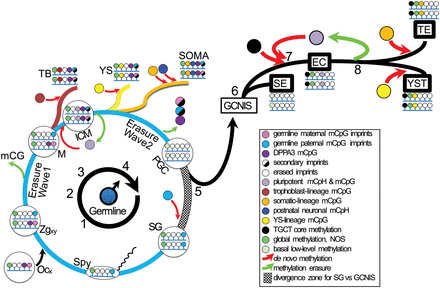
Male germline programming to TGCTs. (1) A sperm with a Chromosome Y (Spy) fertilizes an oocyte (Ocx) to spawn a male zygote (Zgxy) with heteroparental germline imprints. DPPA3 methylation from the egg. (2) Cell divisions and post-zygotic genome-wide CpG methylation (mCpG) erasure en route to the morula (M), with persistent imprints and DPPA3 methylation. (3) Morula progression to blastocyst, with lineage-respective methylation of trophoblast (TB) and inner cell mass (ICM). Prominent pluripotency-signifying methyl-CpH (mCpH-pluri) in ICM stem cells is lost as they exit from pluripotency as yolk sac (YS) and somatic (SOMA) derivatives. Secondary imprints appear in TB and ICM. Differentiation-coupled mCpG, and post-natal neuron-specific CpH (mCpH-neuro). (4) Germline-specific near-total erasure wave eliminates imprints and DPPA3 methylation in the primordial germ cell (PGC) state. (5) PGCs either proceed to spermatogonia (germ cells of later spermatogenesis) with acquisition of paternal imprints and continuation of the physiologic cycle, or they pathologically exit the cycle to be first recognized as germ cell neoplasia in situ (GCNIS) within testicular seminiferous tubules. (6) GCNISs progress to invasive TGCT along two major branches. Seminoma (SEs) are nullipotent and propagate the erased genome of PGCs. (7) De novo methylation of a malignancy program and mCpH-pluri accompany reversion to pluripotency and characterize embryonal carcinoma (EC). (8) EC differentiation to teratoma (TE) or yolk sac tumor (YST) entails loss of mCpH-pluri and convergent reprogramming toward physiologic ICM differentiated derivatives. Global methylation not otherwise specified (NOS) may be found throughout, except in PGCs and SEs, which nevertheless have detectable basal methylation levels. For abbreviations used, see text and legend above.











