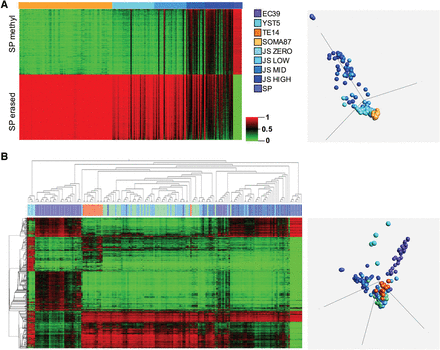
Benign neighboring testis (BNT) methylation profile is determined by spermatogenic proficiency. (A, left) Unclustered heatmap of BNT annotated for Johnsen score (JS) of zero (n = 39), low (n = 30), intermediate (n = 17), or high (n = 27), with reference sperm (SP, n = 8) and somatic tissues/cultures (SOMA, n = 87). 450K array methylation variables were preselected for the top 1000 hyper- and hypomethylated targets in SP relative to SOMA by fold-change (SP erased: SOMA > SP fold-change = 1.915, n = 998 targets; SP methylated: SP > SOMA fold-change = 1.685, n = 998 targets). (Right) PCA on same targets. BNT methylome resembles a two-component mixture of SP and SOMA, with proximity to SP versus SOMA correlated to JS. (B, left) 2D hierarchical clustering of NSE and BNT samples and DMT2000 variables. Note clustering of TE with zero- and low-JS BNT and of high-JS BNT with SP. (Right) PCA of same. NSEs are orthogonally methylated to SPs. For abbreviations used, see text.











