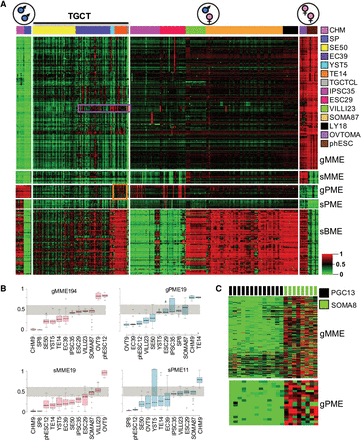
Analysis of imprint erasure in TGCT subtypes. (A) 2D heatmap of imprinted target methylation in TGCTs and references. The heatmap conveys general erasure of maternal and paternal germline (gMME, gPME) and secondary/somatic (sMME, sPME) imprinted methylation, along with differentiation-associated biallelic somatic de novo methylation (sBME), particularly evident in TEs. Like ESCs and TGCTs in vitro, TGCTs show focalized recurrent hypermethylation affecting imprinted loci, such as HM13 in NSEs and H19 in TEs (purple and orange boxes, respectively). (B) Box plots of imprint erasure in TGCT subtypes following adjustment to account for lymphoid infiltrates (see Methods). gMMEs are erased in all subtypes; interestingly, gPMEs are erased in three of four TGCT subtypes, while as noted in A, TEs manifest hypermethylation of H19. Both sMMEs and sPMEs are erased in TGCT subtypes, including TEs. (Nonadjusted measures are provided in Supplemental Fig. S12.) (C) Reference WGBS data show gMME and gPME imprint erasure in PGCs, with maintenance in fetal soma. For abbreviations used, see text.











