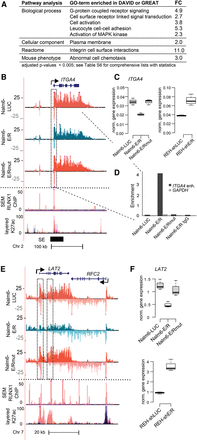
Transmembrane signaling is affected by E/R. (A) Excerpt of the most significantly enriched gene ontology and pathway terms for E/R-regulated transcript-centric and enhancer-centric analyses (see also Supplemental Table S6). (B) GRO-seq and ChIP-seq signals at the ITGA4 locus are shown as in Figure 3E. The annotated SE region present in both CD19+/CD20+ cells and CD34+ cells (Hnisz et al. 2013) harbors a prominent RUNX1 peak downstream from the TSS. This RUNX1 peak is referred to in D. The primary transcript is repressed by E/R. (C) ITGA4 mRNA expression level as measured by RT-qPCR after 24 h of E/R induction (Nalm6-E/R) or after silencing of endogenous E/R in REH cells (REH-shE/R). A representative experiment with technical variation (1.5 × IQR) is shown. Expression is normalized to the housekeeping gene GAPDH. (D) ETV6 ChIP assay with qPCR (primer sites are in the middle of the RUNX1 peak highlighted in B) validates the binding of E/R after 24 h induction, while E/Rmut, LUC, or nonspecific IgG antibody shows no enrichment in comparison to the control region at the GAPDH promoter area. A representative figure of two independent ChIP experiments is shown. (E) LAT2 transcript is repressed in Nalm6-E/R cells (refer to Fig. 3E for tracks). Dashed boxes indicate the TSS and intron 3 of LAT2. (F) Expression of LAT2 mRNA relative to housekeeping gene HMBS as measured by RT-qPCR after induction of E/R for 24 h or after silencing of endogenous E/R in REH cells. A representative experiment is shown with technical variation (1.5 × IQR).











