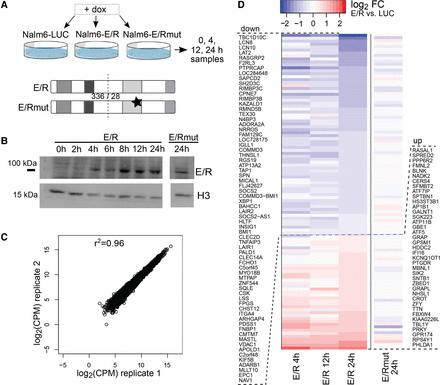
An inducible cell culture model uncovers early transcriptional changes downstream from the ETV6-RUNX1 fusion protein. (A) The doxycycline-inducible cell culture model (Nalm6-E/R) with two controls (Nalm6-E/Rmut and Nalm6-LUC) is schematically illustrated and the sampling time points indicated (see also Supplemental Fig. S1). Mutation at the DNA-binding domain of E/R is marked with a star. (B) Expression of E/R fusion protein after doxycycline induction at indicated time points (a representative Western blot of two replicates). The H3 antibody was used as a loading control. (C) Highly consistent results were obtained from biological replicates, as shown by a correlation plot depicting the GRO-seq signal from Nalm6-LUC cells. Pearson r2 values for the biological replicate pairs of all samples were between 0.96–0.98. (D) A heatmap illustrating magnitude and direction of changes in the GRO-seq signal for the annotated transcripts altered in the E/R sample (and not in E/Rmut) at 24 h. Log2 fold changes of indicated samples relative to Nalm6-LUC are shown in color with shades of blue and red indicating down-regulation and up-regulation, respectively. E/R-mediated changes were predominantly repressive (for genes, see Supplemental Table S5).











