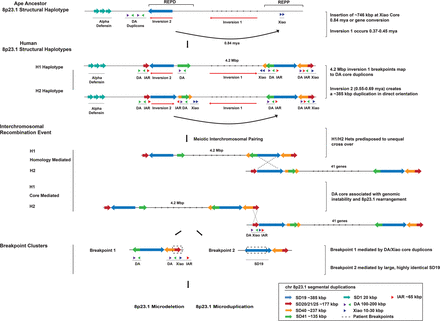
Chromosome 8p23.1 evolutionary and instability model. The model shows the evolution and organization of SDs at Chromosome 8p23.1. Colored arrows represent the largest and most highly identical SDs spanning the critical region. At the top, a schematic of the likely primate ancestral organization of the region based on the sequence assembly of the orangutan REPD and REPP units excluding lineage-specific expansions. A ∼746-kbp duplicative transposition or gene conversion to the proximal side of REPP occurred ∼0.84 mya ± 0.99 (see Supplemental Section 4.2 for timing estimate) and inserted into the Xiao core duplicon at REPP. Two large-scale inversions led to the formation of H2 and H1 in the human lineage (0.5–0.6 mya) with breakpoints associated with the DA/Xiao core duplicons. The H1/H2 configuration promotes interchromosomal nonallelic homologous recombination (NAHR) between directly orientated SDs (SD19, blue arrow) that flank the disease-critical region. The size (385 kbp) and large number of tracts of perfect sequence identity suggest that these rearrangements are driven by NAHR in a subset of patients (n = 3). The second group of patients (n = 4) shows breakpoints mapping to a second pair of duplications (SD20-25, red arrow) corresponding to the DA/Xiao core duplicon. The pair is smaller with only three tracks of perfect sequence identity (>500 bp). We propose that group 2 patients arise as a result of recombination between DA/Xiao core instability elements. Colored arrows represent the length and orientation of SD pairs on each haplotype >100 kbp. The positions of the DA, Xiao, and IAR elements are indicated beneath the SDs (green, purple, blue, and red triangles).











