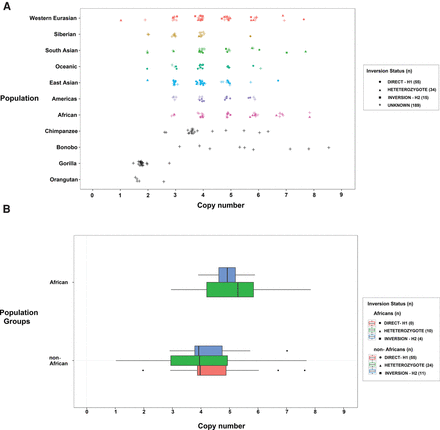
Copy number diversity of the beta-defensin cluster. (A) Copy number estimates for the beta-defensin segmental duplication (SD19) based on sequence read depth from 236 humans and 56 great ape genomes. Copy number represents the diploid aggregate (combined proximal and distal beta-defensin segments at REPP and REPD). (B) Box plots show that Africans have a significantly higher mean beta-defensin copy number compared to non-Africans (P = 0.012). No copy number difference is observed between homozygous direct (H1/H1) versus inverted (H2/H2) haplotypes (P = 0.854). Nonhuman great apes show a copy number consistent with no duplication (diploid copy number 2), with the exception of chimpanzee and bonobo where independent expansions of the beta defensins are predicted to have occurred. The less discrete diplotypes in great apes (i.e., noninteger) are likely a reflection of fewer reads mapping between ape and human reference sequences.











