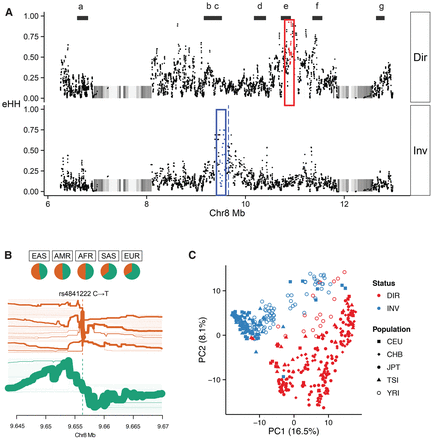
Genetic diversity within the 8p23 inversion. (A) Patterns of extended haplotype homozygosity (eHH) (each point = 20 SNV window) are shown for homozygous direct (H1/H1) and inverted (H2/H2) haplotypes from HGDP samples (Sudmant et al. 2015a). Two regions of eHH are highlighted, eHHI (red box) and eHHD (blue box), with the maximal eHH SNP windows (red and blue dots) and a SNP (rs4841222, blue dashed line) commonly associated with the inverted haplotype. Black bars (a–g) represent haplotypes that were fully resolved by sequencing fosmids from seven individuals (Supplemental Table 14). (B) A bifurcation diagram depicting haplotype sharing from rs4841222 (a marker for the inverted H2 haplotype) as a function of genomic distance from Phase III 1000 Genomes Project. The green bifurcation diagram depicts extensive haplotype sharing consistent with linkage to the H2 haplotype distribution. The frequency of rs4841222 (T) is 0.763 for the PFIDO inverted haplotype and 0.374 for the PFIDO direct haplotype. (C) A principal component analysis for the 1000 Genomes Project for the eHHI block shows almost complete discrimination of the haplotypes in that region irrespective of the human population (shapes). The color denotes the inversion status.











