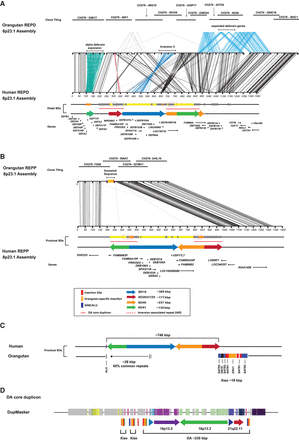
Orangutan sequence assembly of REPD and REPP orthologous regions. (A) A 1.56-Mbp region in the orangutan orthologous to REPD in humans was sequenced and assembled from 10 finished BAC clones and compared using Miropeats (see Fig. 1, H2 for annotation). Lines connecting the two sequences indicate regions of homology and line colors highlight differences between the orangutan and human contig. With the exception of inversion 2, which is in direct orientation (dark blue), the orangutan locus is orientated similarly to human H2. It is >200 kbp larger than human REPD, in part due to a 80-kbp expansion of the alpha-defensin cluster (aqua) and two lineage-specific expansions of beta-defensin (DEFB130 to DEFB136) genes (light blue). (B) A 615-kbp region orthologous to REPP was sequenced and assembled from four finished BAC clones. Comparison with the human H2 assembly using Miropeats shows that the orangutan is missing almost all the duplications (∼746 kbp) present on the human proximal H2 haplotype, including the beta-defensin gene cluster. Black lines connect regions of unique sequence that are shared between orthologous regions in the orangutan and human assemblies. An additional ∼28.8 kbp of ancestral repeat-rich sequence (>65% common repeats) is identified in the orangutan (orange box). (C) A schematic of the proximal insertion site is shown comparing human and orangutan. Sequence analysis of the breakpoint region shows an AluS element mapping on the telomeric side of the 746-kbp duplication block, which is flanked by the 28.8 kbp of ancestral sequence that was lost from the primate ancestor. The centromeric breakpoint maps to a ∼18-kbp core duplicon (Xiao element) that contains OR7E, a gene flanked by SATR2, SATR1, and ERV1 repeat sequences. (D) A schematic description of the DA core duplicon. DupMasker annotation maps the ancestral duplicons to Chr 16p13.3 and Chr 21q22.11; these duplicons form part of a larger complex duplication block referred to as DA. The Xiao core duplicon demarcates the integration site of the SD and is part of the larger DA composite repeat which defines the inversion-associated breakpoint sequences.











