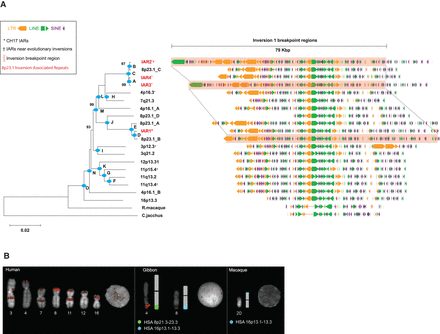
Phylogenetic and comparative analysis of IAR core duplicons. (A) Phylogeny of the core duplicon based on an unrooted neighbor-joining tree (MEGA5) from a 9.2-kbp MSA constructed from 21 IAR core sequences obtained from human and nonhuman primate genome assemblies. Allelic H2 IARs overlapping 8p23.1 inversion breakpoint regions are highlighted in red. The phylogeny shows that the core originated from 16p13.3 and subsequently expanded to various chromosomes early in the ape lineage after divergence from the Old World monkey (see Supplemental Table 10 for timing estimates for nodes A–O, and bootstrap support is indicated). Repeat analysis of a 79-kbp segment associated with the inversion (highlighted) shows that 80% of the structure consists of various classes of common repeats, including LINEs (green), LTRs (orange), and SINEs (purple). (B) FISH analysis using a probe containing core IAR sequences from the gibbon BAC library (CH271-9G12) for human, gibbon (Nomascus leucogenys), and macaque (Macaca mulatta) chromosomal metaphase spreads. Gibbon signals are observed on Chromosomes 4 and 8, which are partially homologous to human Chromosomes 8 (8p21.3–23.3, displayed in light green) and 16 (16p13.1–13.3, displayed in light blue). Macaque signals on Chromosome 20 are homologous to human Chromosome 16p13.3.











