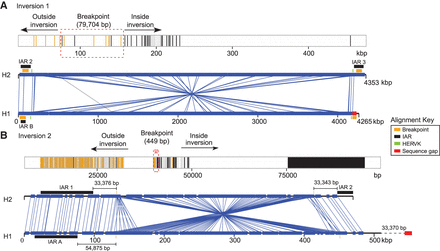
Sequence refinement of 8p23.1 inversion breakpoints within homologous sequences. (A) A 400-kbp multiple sequence alignment (MSA) of homologous sequence between H1 and H2 haplotypes (GRCh37 Chr 8: 7886497–8286302 and Chr 8: 11993961–12171854) was used to refine the breakpoint of inversion 1. Sequence differences mapping inside (black) and outside (yellow) the inversion are depicted in the MSA (top panel). Regions of perfect sequence identity >100 bp are highlighted in dark gray. Breakpoint intervals (dashed red box) are refined (transition region from yellow to black) to a 79.7-kbp region contained within IAR2 and IAR3. The 4.2-Mbp inversion is depicted using Miropeats (lower panel) (GRCh37 Chr 8: 7906497–12171854) with the relative positions of HERVK, IAR, and sequence gaps indicated. The presence of a sequence gap in the H1 assembly (red) prevents further refinement of the inversion 1 breakpoint. (B) Similarly, a schematic of a 100-kbp MSA shows a transition region from yellow to black lines for inversion 2 (GRCh37 Chr 8: 7090000–7175000 and Chr 8: 7370000–7474000). It localizes to a 449-bp region within 33 kbp of IARs 1 and 2. The availability of complete sequence (GRCh37 Chr 8: 6990000–7500000) from H1 and H2 haplotypes allows fine-scale resolution of the inversion 2 breakpoint.











