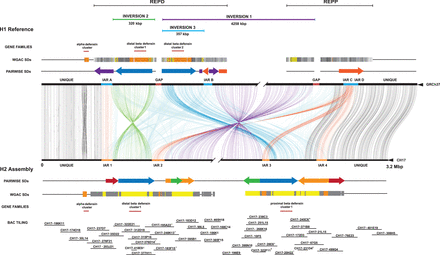
Structural differences between the reference and the inverted 8p23.1 human haplotypes. The human reference genome sequence (GRCh37 representing the direct haplotype H1) is compared to the inverted haplotype (H2 alternate reference assembly) using Miropeats (Parsons 1995). Joining lines indicate homologous sequence between the two haplotypes with the extent and orientation of segmental duplications (SDs) shown in the context of REPD (GRCh37 Chr 8: 6839961–8100062) and REPP (GRCh37 Chr 8: 11860821–12573597). Colored arrows represent the size and orientation of pairwise SDs >100 kbp. The H2 haplotype contains the largest duplication in direct orientation (385 kbp) (SD19 colored blue) creating susceptibility to unequal crossover. Colored bars represent duplication blocks defined using whole-genome assembly comparison (WGAC). These duplication blocks mediate recurrent microdeletion associated with disease. Three large-scale inversions are depicted: two polymorphisms including the large 4.2-Mbp inversion 1 (purple) and a smaller (∼320 kbp) inversion 2 (green) as well as an artifact of the GRCh37 assembly (inversion 3 indicated in blue). A core duplicon (orange bar) defines the inversion breakpoints and are indicated here as inversion-associated repeats (IARs). The locations of alpha- and beta-defensin copies as well as the tiling path of clones sequenced and assembled are depicted. Sequenced clones that could not be fully resolved due to collapse of large internal duplications are highlighted with an asterisk. Clones confirmed to contain small inserts based on fingerprinting are annotated by a dagger. Redundant clones in the tiling path that were sequenced but incompletely assembled are designated with an x. Although a complete 6.3-Mbp assembly was generated, the schematic focuses on SD blocks contained within the flanking sequence of the H2 assembly (breaks in the unique sequence are shown). The higher-level sequence organization of the new assembly was confirmed by a BioNano Genomics map and BAC-end sequence mapping.











