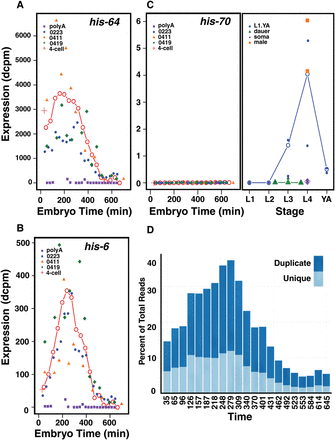
Histone expression across development. (A) The expression in dcpm of the replicative histone, his-64, an example of a pattern with a substantial maternal component. The expression is shown across embryogenesis (labeled in embryo time) for the unified series (red line, open circles), the three rRNA subtracted series (blue, orange, and green), and the poly(A)-selected series (purple) along with the single four-cell sample (red cross). Its pattern is typical of the genes located in the clusters on Chromosome IV. (B) The expression in dcpm of the replicative histone, his-6, an example of a largely zygotically expressed histone. The pattern is shown across the same time points, colored as in A. Its expression pattern is typical of the histone clusters on Chromosomes I and V. (C) The expression in dcpm of his-70, a C-terminally truncated H3.3 variant (Ooi et al. 2006), is shown across embryogenesis (left) and into larval and adult time points (right). The gene lacks an intron but apparently has a poly(A) tail. The embryogenesis time points are colored as in A, the late time points are for L1 through young adult (blue), the dauer stages (green), soma (purple), and male (orange). Both the individual samples (closed symbols) and weighted averages (open symbols) are shown. Post-embryonic time points are shown in approximate chronological order. (D) The percentage of total aligned sequence reads specific to histone genes during embryonic development (time in minutes) for the 0223 rRNA subtracted series (0223) is shown. Uniquely aligned reads are shown in light blue, whereas sequence reads aligning equally to multiple histone gene family members are shown in dark blue.











