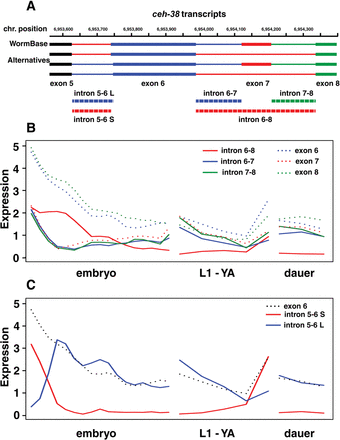
Alternative splicing in exons 5–9 of the transcription factor gene ceh-38. (A) In WormBase, only the topmost gene model is represented, using introns 5-6 S, 6-7, and 7-8 as illustrated. Our data show the presence of additional introns 5-6 L and 6-8. The former deletes two amino acids from exon 6, changing the spacing between the cut (in exon 5) and homebox (in exon 6) DNA binding domains. Intron 6-8 skips exon 7, but maintains the reading frame. (B) Expression data in dcpm for introns 6-7, 7-8, and 6-8 as well as exons 6, 7, and 8 indicate that although the included form is maternally inherited and then lost rapidly, the skipped form is expressed in the early zygote as well as maternally inherited. (C) Expression data in dcpm for introns 5-6 S and 5-6 L show that the shorter intron is maternally inherited and degraded rapidly. In contrast, the longer form has little maternal contribution, rises rapidly in the early embryo, and persists into the later embryo and larval stages, albeit at lower levels.











