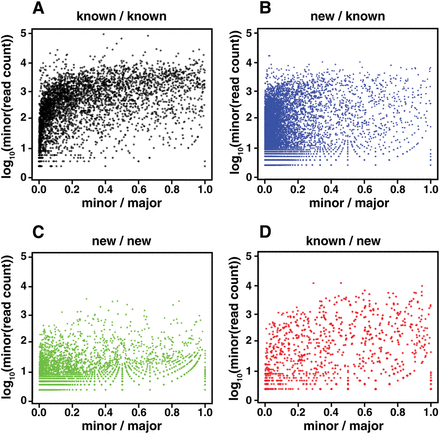
The relative abundance of novel versus known junctions in alternatively spliced pairs within WormBase gene models. For each splice junction site that could be spliced to two or more other sites, i.e., alternatively spliced, we calculated the ratio of reads for each minor form versus the major form and the numbers of reads spanning the minor form. (A) Sites where both the major and minor isoforms were both present in WormBase. (B) Sites where the major isoform was present in WormBase but the minor form was not. (C) Sites where neither the major nor minor isoform were present in WormBase. (D) Sites where the minor form was present in WormBase, but not the major isoform. For the case in which the minor form is novel and the major form is known (B), a larger fraction of the minor form junctions are rare, e.g., ≤100 reads and ≤5% of the major form, than in the case in which both forms are known (A). However, the absolute number of pairs in which the minor form is not rare is almost twice the number of junctions annotated in WormBase (6782 versus 3428), and the other two cases (C,D) add another 4527 relatively well represented alternatively spliced junctions not in WormBase. The rare junctions could represent splicing errors, but considering their overlap in representation with junctions annotated in WormBase, they could also be biologically important.











