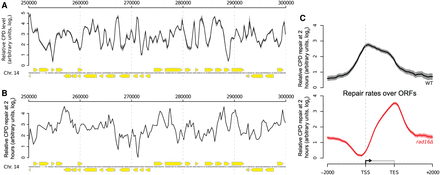
Genome-wide UV-induced DNA repair is organized around gene structure. (A) A linear genome plot of a section of Chromosome 14 showing 3D-DIP-Chip results from wild-type cells. The black line shows the mean (n = 3) CPD level observed immediately after UV irradiation (100 J/m2, shading highlights the SEM). Gray dots indicate the positions of microarray probes. Yellow arrows indicate ORF positions and their direction of transcription. CPD levels are plotted as arbitrary units on the y-axis. (B) CPD repair rates displayed in a linear genome plot. The black line shows the mean of CPD levels 120 min post-UV (n = 2) subtracted from the mean at 0 min post-UV shown in A. Annotations are as described in A. (C) Relative rates of CPD repair around ORF structures. Solid lines show the mean of CPD repair rates in wild-type (n = 3, black line) and rad16Δ cells (n = 2, red line). Shaded areas indicate the SD, with CPD levels plotted as arbitrary units on the y-axis.











