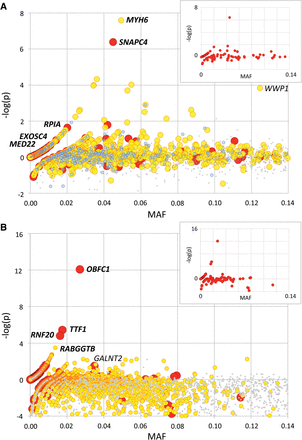
Statistical significance [−log(p): y-axis] of the depletion (positive values) or excess (negative values) in homozygotes for loss-of-function (red; defined as frame-shift, splice-site, and stop-gain variants), missense (yellow), matched synonymous (blue), and random neutral (small gray) variants ordered by minor allele frequency (MAF: x-axis), based on the genotyping of 6385 healthy BBC (A) and 35,219 healthy NZDC (B) animals. Variants that have been subsequently tested in carrier × carrier matings and proven to be embryonic lethals (EL) are labeled in italics and bold. WWP1, shown to affect muscularity, and GALNT2, shown to cause growth retardation, are labeled in italics. For NZDC (B), MAFs were computed across breeds (NZ Holstein-Friesian, NZ Jersey, and NZ cross-bred), explaining the differences with the within-breed MAF reported in Table 2, and the high proportion of variants with negative −log(p) values. Insets: loss-of-function-variants-alone graphs for the corresponding BBC (A) and NZDC (B) populations.











