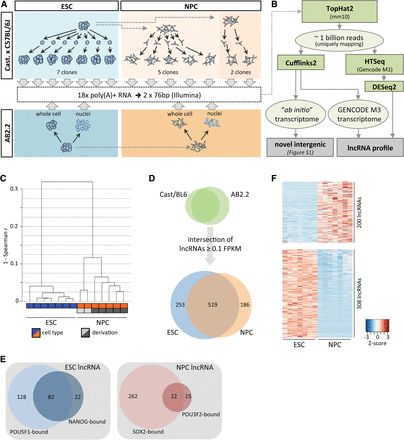
Transcriptome characterization in ESCs and NPCs. (A) Schematic of RNA sources for high-throughput RNA sequencing. (B) Pipeline for ESC and NPC transcriptome analysis. (C) Average linkage clustering dendrogram based on Spearman rank correlation of mRNA and lncRNA expression levels. (D) LncRNAs in the intersection of AB2.2 and Cast/BL6 backgrounds detected in ESCs and NPCs. (E) Analysis of POU5F1, NANOG, SOX2, and POU3F2 binding within 2.5 kb of the transcription start site of expressed lncRNAs (ChIP-seq data reanalyzed from Marson et al. 2008; Lodato et al. 2013). (F) Heatmap (gene-wise Z-score) of lncRNAs in Cast/BL6 clones determined as differentially expressed between ESCs and NPCs (FDR < 0.01).











