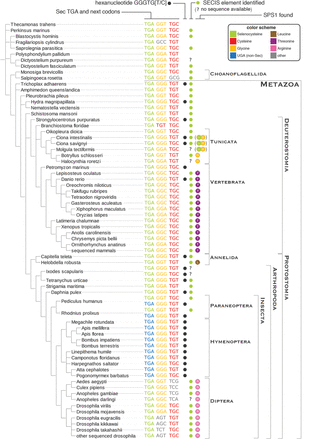
Readthrough-enhancing hexanucleotide in SPS genes. The phylogenetic tree on the left shows the nucleotide sequence alignment at the UGA (or homologous) site in SPS sequences. Only SPS2 and SPS1-UGA genes are shown here. Codons are colored according to their translation, following the same color schema used in Figures 2 and 4 (gray for other amino acids). The presence of the hexanucleotide described in Harrell et al. (2002) is marked with a black dot. Green dots mark the genes for which a bona fide SECIS element was identified. The last column indicates the presence of SPS1 genes.











