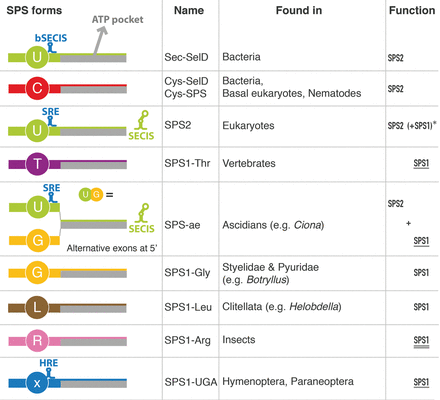
Structure and function of the identified SPS genes. SPS proteins are classified according to the residue found at the UGA or homologous position (Fig. 3). The presence of specific secondary structures is also indicated: (bSECIS) bacterial SECIS element; (SRE) Sec recoding element (Howard et al. 2005); (SECIS) eukaryotic SECIS element; (HRE) hymenopteran readthrough element. The rightmost column indicates the functions predicted for the SPS proteins. SPS2 function is the synthesis of selenophosphate. SPS1 function is defined as the uncharacterized molecular function of Drosophila SPS1-Arg (double underlined), which is likely to be similar to that of other SPS1 genes, as suggested by knockout-rescue experiments in Drosophila (underlined). (*) Eukaryotic SPS2; the parentheses indicate that some such genes are predicted to possess both SPS1 and SPS2 functions, those marked also with a star (*) in Figure 3 (essentially all metazoans with no SPS1 protein in the same genome).











