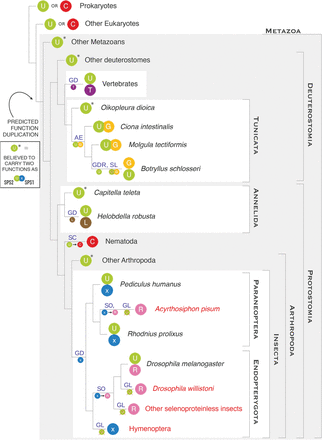
Parallel gene duplications of SPS proteins in metazoa. The plot summarizes the phylogenetic history of metazoan SPS genes, consisting of parallel and convergent events of gene duplication followed by subfunctionalization. Each colored ball represents a SPS gene, indicating the residue found at the UGA or homologous codon: (U) selenocysteine; (C) cysteine; (T) threonine; (G) glycine; (L) leucine; (R) arginine; (x) unknown residue. The gene structures are schematically displayed in Figure 4. The names of the insect species lacking selenoproteins are in red. The main genomic events shaping SPS genes are indicated on the branches: (GD) whole gene duplication; (GDR) gene duplication by retrotransposition; (AE) origin of an alternative exon; (SL) Sec loss; (SC) conversion of Sec to Cys; (SO) conversion of Sec to something other than Cys; (GL) gene loss. In our subfunctionalization hypothesis (see text), we map the origin of a dual function at the root of metazoa. A star (*) marks the metazoan SPS2-Sec genes which did not duplicate. These genes are expected to possess dual function.











