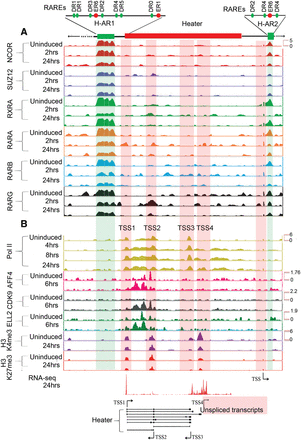
Analysis of changes in the epigenetic state and retinoid receptor occupancy in and around the Heater region during RA-induced differentiation. (A) Occupancy of retinoic acid receptors (RXRA, RARA, RARB, and RARG) and the NCOR corepressor in and around the Heater region. ChIP-on-chip analysis shows a large 2.5-kb region (H-AR1) bound by RAR/RXRs and NCOR upstream of Heater and a smaller domain (H-AR2) region downstream from uninduced and RA-treated ES cells. The schematic at the top shows the relative positions of these regions to Heater and the relative positions of predicted consensus Direct Repeat motifs recognized by retinoid receptors (see Supplemental Fig. S7). (B) Dynamic occupancy of Pol II and components of an elongation complex in the Heater region. H3K27me3 is used as a repressive mark, H3K4me3 is used as a mark for promoters and active chromatin state, and Pol II is used as a mark of active transcription. Along with Pol II, a bivalent mark formed by H3K4me3 and H3K27me3 is noticeable over TSS1-TSS4. There is a rapid recruitment of the transcription elongation factors (ELL2, AFF4, and CDK9) upon RA induction. Tracks were configured by using a windowing function as mean and smoothing windows as 10 pixels. At the bottom, multiple Heater transcripts are shown schematically with the respective transcription start sites (TSS) along with RNA-seq at 24 h. The direction of transcription is indicated by arrows. The pink box denotes a region with a large number of unspliced transcripts generated from TSS4. A putative TSS is also apparent in the H-AR2 region.











