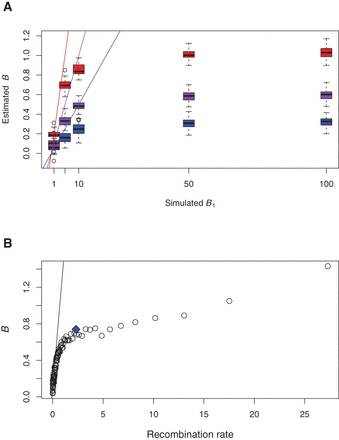
Effect of hotspots on B estimates. (A) Simulations were performed under model M2b with B0 = 0, f = 0.05 (blue), 0.1 (purple), and 0.2 (red). For each B1 value (x-axis), 100 simulations were performed and the M1 model was applied to estimate B. The lines correspond to the expectation B = (1 − f)B0 + fB1. Very similar results were obtained for B0 = 0.25, and B0 = 0.5 (not shown). (B) The whole data set was divided into centiles of recombination rates computed over 5 kb around each SNP. The line corresponds to the regression performed on centiles for which the recombination rate was lower than 0.1 cM/Mb. Dots correspond to B estimates under the M1* model. The blue diamond corresponds to B estimated using the SNPs belonging to the gBGC tracts detected by Capra et al. (2013).











