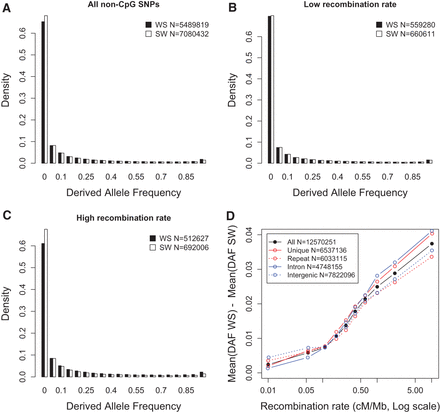
Variations in derived allele frequencies (DAF) according to mutation type (WS or SW) and local recombination rate: non-CpG sites. SNP allele frequencies and polarizations were retrieved from the 1000 Genomes phase 1 data set (population panel: AFR). We selected all non-CpG SNPs located in noncoding regions from autosomes. Recombination rates are measured over 5-kb windows centered on each SNP, using HapMap data (Myers et al. 2005). (A) DAF spectra of all SNPs. (B) DAF spectra of the subset of SNPs located in regions of low recombination (bottom 10%). (C) DAF spectra of the subset of SNPs located in regions of high recombination (top 10%). (D) Differences in mean DAF spectra between WS and SW mutations, according to the local recombination rate. These differences are displayed for the entire set of SNPs, for the subsets of SNPs located in introns versus intergenic regions (blue) or in unique versus repeat sequences (red). The values in the insets indicate the genome-wide count of SNPs of each category.











