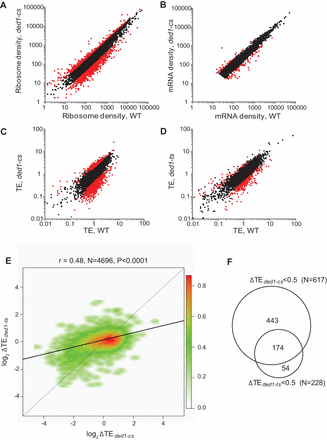
ded1-cs and ded1-ts mutations evoke genome-wide changes in relative TE values for substantial fractions of genes that overlap significantly. (A,B) Scatterplots of normalized read densities expressed as number of reads mapping to gene coding sequences normalized by coding sequence length and by total number of reads for that sample. (A) Ribosome footprint density and (B) mRNA density for ded1-cs (NSY5) versus WT (NSY4) strains. (C) Scatterplot of translational efficiencies (TEs) between WT and ded1-cs. Eight-hundred fourteen genes exhibiting ≥twofold changes in TE in ded1-cs cells at FDR < 0.01 are in red. (D) Same as C for WT (NSY10) versus ded1-ts (NSY11) strains; 336 genes with ≥twofold change in TE in ded1-ts cells at FDR < 0.01 colored red. (E) Smooth scatterplot of log2ΔTE values for 4696 expressed genes for ded1-ts versus ded1-cs strains, excluding genes with log2ΔTE >6 or <−6. Black line: determined regression line; gray line: theoretical regression line for identical changes in ΔTE values. Plot was generated using R smoothscatter function with 128 color bins and kernel density estimate. The red to green gradient indicates density of data points. (F) Venn diagram of overlap between genes exhibiting ≥twofold reductions in TE in the two ded1 mutants.











