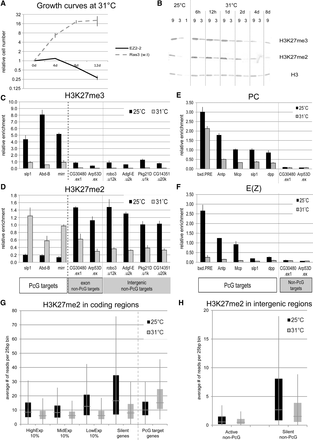
Global changes of H3K27me2/3 levels after E(z) inactivation. (A) Growth curves of EZ2-2 and Ras3 (w.t.) cells after shift to 31°C. (B) Global H3K27me2/3 levels in EZ2-2 cells at 25°C and 31°C assayed by Western blot. Threefold serial dilutions of nuclear extract were loaded. Total H3 is the loading control. (C,D) H3K27me3 and H3K27me2 levels at PcG target and non-PcG target regions from EZ2-2 cells at 25°C and 31°C. H3K27me2/3 levels were measured by ChIP-qPCR from at least two independent experiments. The relative enrichment values were calculated based on the qPCR values normalized to input DNA. Error bars, SE. The four intergenic regions are identified by the distance in kb from the TSS (e.g., u12k). (E,F) The levels of Pc and E(z) at PcG target regions and two non-PcG target regions assayed by ChIP-qPCR from EZ2-2 cells grown at 25°C and 31°C. (G) H3K27me2 levels in highly expressed (HighExp 10%), intermediate (MidExp 10%), weakly expressed (LowExp 10%), and silent (no RNA-seq reads) non-PcG target genes and PcG target genes were analyzed from EZ2-2 H3K27me2 ChIP-seq data at 25°C and 31°C. (H) H3K27me2 levels in transcriptionally active intergenic non-PcG target (Top 10%) and silent regions (no RNA-seq reads) were analyzed from EZ2-2 H3K27me2 ChIP-seq data at 25°C and 31°C.











