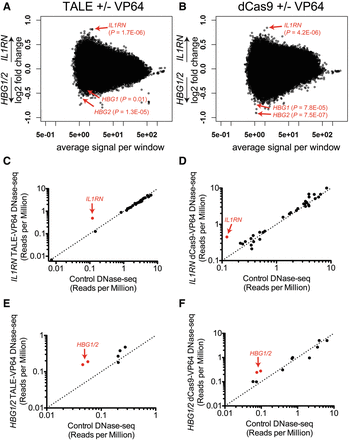
Global characterization of changes to chromatin accessibility. (A) Scatter plot of DNase-seq data comparing samples treated with TALEs, with and without VP64, targeted to IL1RN versus HBG1/2. Each dot represents a DNase I hypersensitive site analyzed by DESeq. IL1RN and HBG1/2 display the expected opposite differences in chromatin accessibility. Nominal P-values for each target site are indicated. (B) Similar scatter plot as A, but for DNase-seq data from IL1RN-targeted dCas9 ± VP64 versus HBG1/2-targeted dCas9 ± VP64. The individual comparisons of all eight treatments compared to control are presented in Supplemental Figures 5, 6; and the top 100 differential DHS sites for each treatment are provided in Supplemental Tables 16–23. (C–F) DNase-seq signal for target (red circles) and off-target ChIP-seq sites (black circles). For each off-target ChIP-seq site, normalized DNase-seq signal from IL1RN-targeted TALE-VP64 (C), IL1RN-targeted dCas9-VP64 (D), HBG1/2-targeted TALE-VP64s (E), and HBG1/2-targeted dCas9-VP64 (F) was compared to normalized DNase-seq signal from control cells transfected with empty plasmid.











