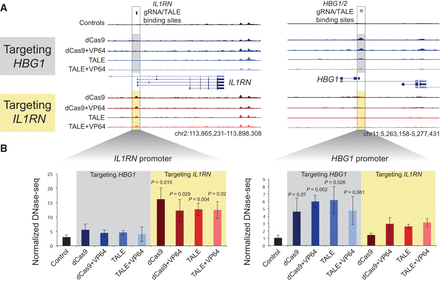
Chromatin accessibility changes induced by both TALEs and dCas9. HEK293T cells were transfected with expression plasmids for TALEs ± VP64 and gRNAs with dCas9 ± VP64 targeted to either the HBG1/2 promoter or the IL1RN promoter. (A) Representative DNase-seq data surrounding each promoter (highlighted in box) show increased chromatin accessibility at the promoter to which the TALEs and dCas9 are targeted, but not at the other promoter. (B) Normalized DNase-seq cut counts within a 300-bp window surrounding each promoter are shown (mean ± SEM, n = 4–6) (Supplemental Table 32). P-values are shown compared to the control sample (Tukey's test).











